Deep Learning Using Histological Images for Gene Mutation Prediction in Lung Cancer: a Multicentre Retrospective Study
Jan 1, 2025·,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,·
0 min read
Yu Zhao*
Shan Xiong*
Qin Ren*
Jun Wang*
Min Li*
Lin Yang
Di Wu
Kejing Tang
Xiaojie Pan
Fengxia Chen
Wenxiang Wang
Shi Jin
Xianling Liu
Gen Lin
Wenxiu Yao
Linbo Cai
Yi Yang
Jixian Liu
Jingxun Wu
Wenfan Fu
Kai Sun
Feng Li
Bo Cheng
Shuting Zhan
Haixuan Wang
Ziwen Yu
Xiwen Liu
Ran Zhong
Huiting Wang
Ping He
Yongmei Zheng
Peng Liang
Longfei Chen
Ting Hou
Junzhou Huang
Bing He
Jiangning Song
Lin Wu
Chengping Hu
Jianxing He
Jianhua Yao
Wenhua Liang
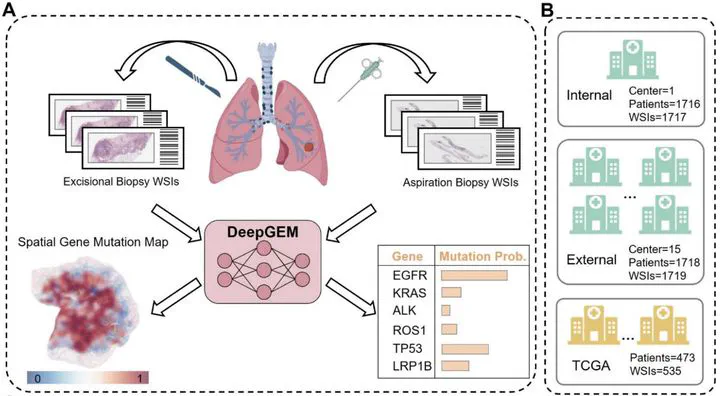 DeepGEM workflow
DeepGEM workflowAbstract
In this multicentre retrospective study, we collected data for patients with lung cancer who had a biopsy and multigene next-generation sequencing done at 16 hospitals in China (with no restrictions on age, sex, or histology type), to form a large multicentre dataset comprising paired pathological image and multiple gene mutation information. We also included patients from The Cancer Genome Atlas (TCGA) publicly available dataset. Our developed model is an instance-level and bag-level co-supervised multiple instance learning method with label disambiguation design. We trained and initially tested the DeepGEM model on the internal dataset (patients from the First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China), and further evaluated it on the external dataset (patients from the remaining 15 centres) and the public TCGA dataset. Additionally, a dataset of patients from the same medical centre as the internal dataset, but without overlap, was used to evaluate the model’s generalisation ability to biopsy samples from lymph node metastases. The primary objective was the performance of the DeepGEM model in predicting gene mutations (area under the curve [AUC] and accuracy) in the four prespecified groups (ie, the hold-out internal test set, multicentre external test set, TCGA set, and lymph node metastases set).
Type
Publication
The Lancet Oncology